Vladislav Varslavans
2018-05-22 11:48:12 UTC
We are currently working on the combination of rstudio-server and conda as
well as we plan to use conda for python package management (*one
environment.yml per Data Science application*). There are a lot of
questions/answers scattered around the internet about this topic, but there
was nowhere completely described how to do it and how to troubleshoot or
solve different troubles that might come up on the way.
We would like to *share our experience* and *hear opinions and critics* for
our approach and rise a discussion on the topic. Also, we would like to
encourage everyone to collect references to other material about the topic
on the question and finally create some complete guide that will help
whoever will want to do the same.
*Environment and overview*
For each user in our system we have independent containers with
rstudio-server. Data Scientists are using both R (and python) for
development. For R weâve used packrat as package manager, but following
concerns arises:
1. Packrat will compile each package first time â that means that
recreation of environment for the project usually takes long time. Also,
creation of clean testing environment is time consuming.
2. Packrat is usable only with R â python developers canât use it - that
means that different projects are using different package managers.
3. Packrat does not handle package dependencies on system libraries (e.g.
RODBC package depends on unixodbc)
4. Packrat cannot handle binary packages that are not available in cran (such
as catboost).
Since conda is language agnostic package and environment manager that
supports number of Rpackages, it gains influence in python world and it has
constantly growing community of contributors who add new packages to conda we
decided to try it out. The solution weâve thought about looks like this:
<Loading Image...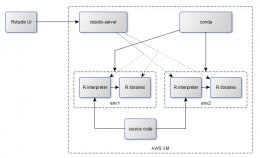 >
>
*The following questions we have formulated*:
1. How to combine conda and the containerized rstudio-server?
2. How to handle packages that are not available in conda?
3. Is it possible to combine conda with install.packages() or
devtools::install_github() for missing packages at least to tryout
packages?
4. Is it possible to combine conda with packrat?
5. How to use conda with spark (sparklyr, pyspark)?
6. Is it feasible to use conda?
*Below are our answers to these questions:*
1. How to combine conda and the containerized rstudio-server?
This is quite simple:
*First* put following line .libPaths(paste0(R.home(), "/library")) inside
~/.Rprofile file. This will exclude paths to any library directory, but the
one that is located in R home directory. This way whenever R home directory
is switched â R library directory will also be changed.
*Second* â substitute line rsession-which-r=path/to/r/home in
/etc/rstudio/rserver.conf file to path to R within conda environment.
Weâve developed following script to do it automatically:
# First argument is a name of the conda environment that needs to be activated
echo Target environment is: $1
# Find path to the environment (second column of output of command)
new_env_path=`conda info --env | grep -E "^$1\s+" | awk '{print $2}'`
# In case if the environment is active in conda - read path in third columnif [ "$new_env_path" = "*" ]; then
echo "Env is active!"
new_env_path=`conda info --env | grep -E "^$1\s+" | awk '{print $3}'`else
echo "Env is not active"
fi;
echo "New path = $new_env_path"
# sudo is required to edit configuraion file
sudo sed -i "s|rsession-which-r=.*|rsession-which-r=$new_env_path/bin/R|" /etc/rstudio/rserver.conf
# ... and to restart service
sudo rstudio-server restart
This will successfully switch the R interpreter and libraries to a selected
conda environment.
2. How to handle packages that are not available in conda?
Here are 2 solution that weâve tried:
*Conda skeleton <https://conda.io/docs/commands/build/conda-skeleton.html>* â
for packages that are available in cran or some other package managers. It
will automatically create and compile package for you and will let you to
(or even automatically) upload the package to anaconda.org own channel. To
use conda skeleton is easy:
conda skeleton cran <package_name>
conda build <package>
The guide on how to use it is here <http://ihrke.github.io/conda.html>.
The error weâve got doing it is:
Undefined Jinja2 variables remain (['cran_mirror', 'cran_mirror']). Please enable source downloading and try again.
This issue can be resolved by creating file with address of the cran mirror:
echo "cran_mirror: https://cloud.r-project.org/" > cfg.yam"
Add argument to build command:
conda build <package> -m cfg.yam
For the packages that are not available in cran itâs possible to use
conda-forge <https://conda-forge.org/> and contribute to community by
creating receipts for the missing packages. The process is well documented
and available here <https://github.com/conda-forge/staged-recipes>.
3. Is it possible to combine conda with install.packages() or
devtools::install_github() for missing packages at least to tryout packages?
It seems that is should be possible for purely R packages, but there are
currently 2 errors with to be compiled packages, as the conda build
environment is not utilized automatically:
1. x86_64-conda_cos6-linux-gnu-cc not found â when trying to compile
package. There is a workaround for it - using Sys.setenv() prepend path
to the conda environment bin folder to PATH variable: Sys.setenv(PATH=paste0("path/to/conda/env/bin:",
Sys.getenv(âPATHâ)))
2. The <somlibrary> library that is required to build <someotherlibrary>
was not found. â There is no currently solution how to fix it, but I
assume, that there should be some complete solution how to fix both, this
and the previous error.
4. Is it possible to combine conda with packrat?
Does not look like as packrat does not recognize packages already installed
via conda and it faces the same build issues as 3.
5. How to use conda with spark (sparklyr, pyspark)?
This question is how to bring conda environment to the executors. Some
similar question was already discussed here
<https://henning.kropponline.de/2016/09/24/running-pyspark-with-conda-env/>,
so thatâs where Iâll start my investigation.
6. Is it feasible to use conda?
*Seems like yes?!* >90-95% of the required R packages are anyway available
via conda-forge. The rest can be handled locally via conda skeleton cran &
build eventually followed by an upload to own anaconda channel or by
contributing to conda-forge. Conda + packrat seems not to work and maybe
also not be required. devtools::install_github() would be nice to have, but
how to get it running if the code needs to be compiled? Is there any other
solution to handle additional packages on top of conda - shell or R setup
script?
well as we plan to use conda for python package management (*one
environment.yml per Data Science application*). There are a lot of
questions/answers scattered around the internet about this topic, but there
was nowhere completely described how to do it and how to troubleshoot or
solve different troubles that might come up on the way.
We would like to *share our experience* and *hear opinions and critics* for
our approach and rise a discussion on the topic. Also, we would like to
encourage everyone to collect references to other material about the topic
on the question and finally create some complete guide that will help
whoever will want to do the same.
*Environment and overview*
For each user in our system we have independent containers with
rstudio-server. Data Scientists are using both R (and python) for
development. For R weâve used packrat as package manager, but following
concerns arises:
1. Packrat will compile each package first time â that means that
recreation of environment for the project usually takes long time. Also,
creation of clean testing environment is time consuming.
2. Packrat is usable only with R â python developers canât use it - that
means that different projects are using different package managers.
3. Packrat does not handle package dependencies on system libraries (e.g.
RODBC package depends on unixodbc)
4. Packrat cannot handle binary packages that are not available in cran (such
as catboost).
Since conda is language agnostic package and environment manager that
supports number of Rpackages, it gains influence in python world and it has
constantly growing community of contributors who add new packages to conda we
decided to try it out. The solution weâve thought about looks like this:
<Loading Image...
*The following questions we have formulated*:
1. How to combine conda and the containerized rstudio-server?
2. How to handle packages that are not available in conda?
3. Is it possible to combine conda with install.packages() or
devtools::install_github() for missing packages at least to tryout
packages?
4. Is it possible to combine conda with packrat?
5. How to use conda with spark (sparklyr, pyspark)?
6. Is it feasible to use conda?
*Below are our answers to these questions:*
1. How to combine conda and the containerized rstudio-server?
This is quite simple:
*First* put following line .libPaths(paste0(R.home(), "/library")) inside
~/.Rprofile file. This will exclude paths to any library directory, but the
one that is located in R home directory. This way whenever R home directory
is switched â R library directory will also be changed.
*Second* â substitute line rsession-which-r=path/to/r/home in
/etc/rstudio/rserver.conf file to path to R within conda environment.
Weâve developed following script to do it automatically:
# First argument is a name of the conda environment that needs to be activated
echo Target environment is: $1
# Find path to the environment (second column of output of command)
new_env_path=`conda info --env | grep -E "^$1\s+" | awk '{print $2}'`
# In case if the environment is active in conda - read path in third columnif [ "$new_env_path" = "*" ]; then
echo "Env is active!"
new_env_path=`conda info --env | grep -E "^$1\s+" | awk '{print $3}'`else
echo "Env is not active"
fi;
echo "New path = $new_env_path"
# sudo is required to edit configuraion file
sudo sed -i "s|rsession-which-r=.*|rsession-which-r=$new_env_path/bin/R|" /etc/rstudio/rserver.conf
# ... and to restart service
sudo rstudio-server restart
This will successfully switch the R interpreter and libraries to a selected
conda environment.
2. How to handle packages that are not available in conda?
Here are 2 solution that weâve tried:
*Conda skeleton <https://conda.io/docs/commands/build/conda-skeleton.html>* â
for packages that are available in cran or some other package managers. It
will automatically create and compile package for you and will let you to
(or even automatically) upload the package to anaconda.org own channel. To
use conda skeleton is easy:
conda skeleton cran <package_name>
conda build <package>
The guide on how to use it is here <http://ihrke.github.io/conda.html>.
The error weâve got doing it is:
Undefined Jinja2 variables remain (['cran_mirror', 'cran_mirror']). Please enable source downloading and try again.
This issue can be resolved by creating file with address of the cran mirror:
echo "cran_mirror: https://cloud.r-project.org/" > cfg.yam"
Add argument to build command:
conda build <package> -m cfg.yam
For the packages that are not available in cran itâs possible to use
conda-forge <https://conda-forge.org/> and contribute to community by
creating receipts for the missing packages. The process is well documented
and available here <https://github.com/conda-forge/staged-recipes>.
3. Is it possible to combine conda with install.packages() or
devtools::install_github() for missing packages at least to tryout packages?
It seems that is should be possible for purely R packages, but there are
currently 2 errors with to be compiled packages, as the conda build
environment is not utilized automatically:
1. x86_64-conda_cos6-linux-gnu-cc not found â when trying to compile
package. There is a workaround for it - using Sys.setenv() prepend path
to the conda environment bin folder to PATH variable: Sys.setenv(PATH=paste0("path/to/conda/env/bin:",
Sys.getenv(âPATHâ)))
2. The <somlibrary> library that is required to build <someotherlibrary>
was not found. â There is no currently solution how to fix it, but I
assume, that there should be some complete solution how to fix both, this
and the previous error.
4. Is it possible to combine conda with packrat?
Does not look like as packrat does not recognize packages already installed
via conda and it faces the same build issues as 3.
5. How to use conda with spark (sparklyr, pyspark)?
This question is how to bring conda environment to the executors. Some
similar question was already discussed here
<https://henning.kropponline.de/2016/09/24/running-pyspark-with-conda-env/>,
so thatâs where Iâll start my investigation.
6. Is it feasible to use conda?
*Seems like yes?!* >90-95% of the required R packages are anyway available
via conda-forge. The rest can be handled locally via conda skeleton cran &
build eventually followed by an upload to own anaconda channel or by
contributing to conda-forge. Conda + packrat seems not to work and maybe
also not be required. devtools::install_github() would be nice to have, but
how to get it running if the code needs to be compiled? Is there any other
solution to handle additional packages on top of conda - shell or R setup
script?
--
You received this message because you are subscribed to the Google Groups "conda - Public" group.
To unsubscribe from this group and stop receiving emails from it, send an email to conda+***@continuum.io.
To post to this group, send email to ***@continuum.io.
Visit this group at https://groups.google.com/a/continuum.io/group/conda/.
To view this discussion on the web visit https://groups.google.com/a/continuum.io/d/msgid/conda/2036a5c1-44ef-462e-b0a0-695a0337c74b%40continuum.io.
For more options, visit https://groups.google.com/a/continuum.io/d/optout.
You received this message because you are subscribed to the Google Groups "conda - Public" group.
To unsubscribe from this group and stop receiving emails from it, send an email to conda+***@continuum.io.
To post to this group, send email to ***@continuum.io.
Visit this group at https://groups.google.com/a/continuum.io/group/conda/.
To view this discussion on the web visit https://groups.google.com/a/continuum.io/d/msgid/conda/2036a5c1-44ef-462e-b0a0-695a0337c74b%40continuum.io.
For more options, visit https://groups.google.com/a/continuum.io/d/optout.